Introduction
Hoea is a python module for hierarchical ontology enrichment analysis, which facilitated GO (Gene Ontology)/ KO (KEGG Orthology) enrichment analysis at any desktop. Enrichment analysis appears to be a routine task for high-throughput experiments such as microarray and RNA-Seq (expression studies based on next generation sequencing data). Hoea aims to make this task easy for bioinformaticans in the dry lab. Hoea supports Gene ontology and KEGG orthology, and is open source. Experienced programmer could easily modified the code to suite for their demands.
Prerequisite
Hoea was written in Python language, statistical calculation was implemented by R, graphs were drew by Graphviz. Please make sure your system was equipped with these tools before using Hoea.
News
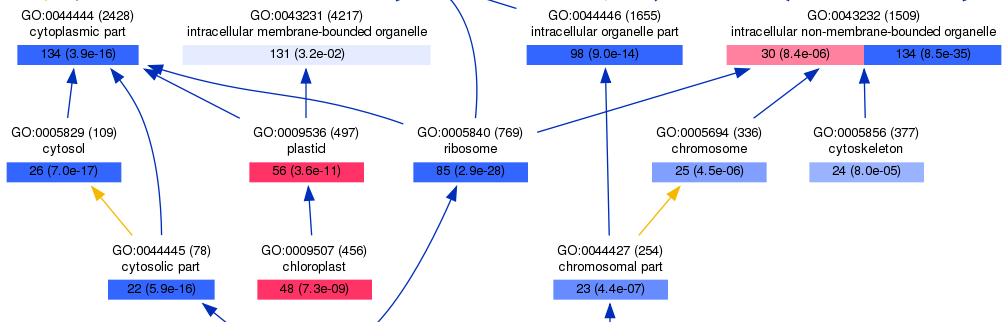
1, New features since version 0.3: Hoea now support two list of input for easier compare analysis. The two list will filled by red and blue serious color, respectively. As illustrated below, the red and blue box indicated up- and down-regulated probes enrichmented GO terms, respectively.
How to use Hoea
Check installation instructions and a quick guide from Tutorial.
Check detail documetation from Documentation.
Contact
Users are encouraged to contact me for suggestions of this module. Any advice will be greatly appreciated.